Introduction: Nitrogen on copper#
This section gives a quick (and incomplete) overview of what ASE can do.
We will calculate the adsorption energy of a nitrogen molecule on a copper surface. This is done by calculating the total energy for the isolated slab and for the isolated molecule. The adsorbate is then added to the slab and relaxed, and the total energy for this composite system is calculated. The adsorption energy is obtained as the sum of the isolated energies minus the energy of the composite system.
Here is a picture of the system after the relaxation:
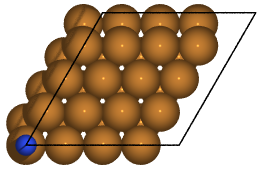
Please have a look at the following script N2Cu.py:
from ase import Atoms
from ase.build import add_adsorbate, fcc111
from ase.calculators.emt import EMT
from ase.constraints import FixAtoms
from ase.optimize import QuasiNewton
h = 1.85
d = 1.10
slab = fcc111('Cu', size=(4, 4, 2), vacuum=10.0)
slab.calc = EMT()
e_slab = slab.get_potential_energy()
molecule = Atoms('2N', positions=[(0.0, 0.0, 0.0), (0.0, 0.0, d)])
molecule.calc = EMT()
e_N2 = molecule.get_potential_energy()
add_adsorbate(slab, molecule, h, 'ontop')
constraint = FixAtoms(mask=[a.symbol != 'N' for a in slab])
slab.set_constraint(constraint)
dyn = QuasiNewton(slab, trajectory='N2Cu.traj')
dyn.run(fmax=0.05)
print('Adsorption energy:', e_slab + e_N2 - slab.get_potential_energy())
Assuming you have ASE setup correctly (Installation) run the script:
python N2Cu.py
Please read below what the script does.
Atoms#
The Atoms object is a collection of atoms. Here
is how to define a N2 molecule by directly specifying the position of
two nitrogen atoms:
>>> from ase import Atoms
>>> d = 1.10
>>> molecule = Atoms('2N', positions=[(0., 0., 0.), (0., 0., d)])
You can also build crystals using, for example, the lattice module
which returns Atoms objects corresponding to
common crystal structures. Let us make a Cu (111) surface:
>>> from ase.build import fcc111
>>> slab = fcc111('Cu', size=(4,4,2), vacuum=10.0)
Adding calculator#
In this overview we use the effective medium theory (EMT) calculator, as it is very fast and hence useful for getting started.
We can attach a calculator to the previously created
Atoms objects:
>>> from ase.calculators.emt import EMT
>>> slab.calc = EMT()
>>> molecule.calc = EMT()
and use it to calculate the total energies for the systems by using
the get_potential_energy() method from the
Atoms class:
>>> e_slab = slab.get_potential_energy()
>>> e_N2 = molecule.get_potential_energy()
Structure relaxation#
Let’s use the QuasiNewton minimizer to optimize the
structure of the N2 molecule adsorbed on the Cu surface. First add the
adsorbate to the Cu slab, for example in the on-top position:
>>> h = 1.85
>>> add_adsorbate(slab, molecule, h, 'ontop')
In order to speed up the relaxation, let us keep the Cu atoms fixed in
the slab by using FixAtoms from the
constraints module. Only the N2 molecule is then allowed
to relax to the equilibrium structure:
>>> from ase.constraints import FixAtoms
>>> constraint = FixAtoms(mask=[a.symbol != 'N' for a in slab])
>>> slab.set_constraint(constraint)
Now attach the QuasiNewton minimizer to the
system and save the trajectory file. Run the minimizer with the
convergence criteria that the force on all atoms should be less than
some fmax:
>>> from ase.optimize import QuasiNewton
>>> dyn = QuasiNewton(slab, trajectory='N2Cu.traj')
>>> dyn.run(fmax=0.05)
Note
The general documentation on structure optimizations contains information about different algorithms, saving the state of an optimizer and other functionality which should be considered when performing expensive relaxations.
Input-output#
Writing the atomic positions to a file is done with the
write() function:
>>> from ase.io import write
>>> write('slab.xyz', slab)
This will write a file in the xyz-format. Possible formats are:
format |
description |
|---|---|
|
Simple xyz-format |
|
Gaussian cube file |
|
Protein data bank file |
|
ASE’s own trajectory format |
|
Python script |
Reading from a file is done like this:
>>> from ase.io import read
>>> slab_from_file = read('slab.xyz')
If the file contains several configurations, the default behavior of
the write() function is to return the last
configuration. However, we can load a specific configuration by
doing:
>>> read('slab.traj') # last configuration
>>> read('slab.traj', -1) # same as above
>>> read('slab.traj', 0) # first configuration
Visualization#
The simplest way to visualize the atoms is the view()
function:
>>> from ase.visualize import view
>>> view(slab)
This will pop up a ase.gui window. Alternative viewers can be used
by specifying the optional keyword viewer=... - use one of
‘ase.gui’, ‘gopenmol’, ‘vmd’, or ‘rasmol’. (Note that these alternative
viewers are not a part of ASE and will need to be installed by the user
separately.) The VMD viewer can take an optional data argument to
show 3D data:
>>> view(slab, viewer='VMD', data=array)
Molecular dynamics#
Let us look at the nitrogen molecule as an example of molecular
dynamics with the VelocityVerlet
algorithm. We first create the VelocityVerlet object giving it the molecule and the time
step for the integration of Newton’s law. We then perform the dynamics
by calling its run() method and
giving it the number of steps to take:
>>> from ase.md.verlet import VelocityVerlet
>>> from ase import units
>>> dyn = VelocityVerlet(molecule, dt=1.0 * units.fs)
>>> for i in range(10):
... pot = molecule.get_potential_energy()
... kin = molecule.get_kinetic_energy()
... print('%2d: %.5f eV, %.5f eV, %.5f eV' % (i, pot + kin, pot, kin))
... dyn.run(steps=20)
