Introduction to GPAW internals¶
This guide will contain graphs showing the relationship between objects that build up a DFT calculation engine.
Hint
Here is a simple graph showing the relations between the classes A,
B and C:
Here, the object of type A has an attribute a of type int and an
attribute b of type B or C, where C inherits from B.
DFT components¶
The components needed for a DFT calculation are created by a “builder” that
can be made with the builder() function, an ASE
ase.Atoms object and some input parameters:
>>> from ase import Atoms
>>> atoms = Atoms('Li', cell=[2, 2, 2], pbc=True)
>>> from gpaw.new.builder import builder
>>> params = {'mode': 'pw', 'kpts': (5, 5, 5)}
>>> b = builder(atoms, params)
As seen in the figure above, there are builders for each of the modes: PW, FD and LCAO (builders for TB and ATOM modes are not shown).
The InputParameters object takes care of
user parameters:
checks for errors
does normalization
handles backwards compatibility and deprecation warnings
Normally, you will not need to create a DFT-components builder yourself. It will happen automatically when you create a DFT-calculation object like this:
>>> from gpaw.new.calculation import DFTCalculation
>>> calculation = DFTCalculation.from_parameters(atoms, params)
or when you create an ASE-calculator interface:
>>> from gpaw.new.ase_interface import GPAW
>>> atoms.calc = GPAW(**params, txt='li.txt')
Full picture¶
The ase.Atoms object has an
gpaw.new.ase_interface.ASECalculator object attached
created with the gpaw.new.ase_interface.GPAW() function:
>>> atoms = Atoms('H2',
... positions=[(0, 0, 0), (0, 0, 0.75)],
... cell=[2, 2, 3],
... pbc=True)
>>> atoms.calc = GPAW(mode='pw', txt='h2.txt')
>>> atoms.calc
ASECalculator(mode: {'name': 'pw'})
The atoms.calc object manages a
gpaw.new.calculation.DFTCalculation object that does the actual work.
When we do this:
>>> e = atoms.get_potential_energy()
the gpaw.new.ase_interface.ASECalculator.get_potential_energy()
method gets called (atoms.calc.get_potential_energy(atoms))
and the following will happen:
create
gpaw.new.calculation.DFTCalculationobject if not already doneupdate positions/unit cell if they have changed
start SCF loop and converge if needed
calculate energy
store a copy of the atoms
DFT-calculation object¶
An instance of the gpaw.new.calculation.DFTCalculation class has
the following attributes:
|
|
|
|
|
and a the gpaw.new.calculation.DFTState object has these attributes:
|
|
|
|
|
Naming convention for arrays¶
Commonly used indices:
index
description
aAtom number
cUnit cell axis-index (0, 1, 2)
vxyz-index (0, 1, 2)
KBZ k-point index
kIBZ k-point index
qIBZ k-point index (local, i.e. it starts at 0 on each processor)
sSpin index (\(\sigma\))
sSymmetry index
uCombined spin and k-point index (local)
RThree indices into the coarse 3D grid
rThree indices into the fine 3D grid
GIndex of plane-wave coefficient (wave function expansion,
ecut)
gIndex of plane-wave coefficient (densities,
2 * ecut)
hIndex of plane-wave coefficient (compensation charges,
8 * ecut)
X
RorG
x
r,gorh
xZero or more extra dimensions
MLCAO orbital index (\(\mu\))
nBand number
nPrincipal quantum number
lAngular momentum quantum number (s, p, d, …)
mMagnetic quantum number (0, 1, …, 2*`ell` - 1)
L
landm(L = l**2 + m)
jValence orbital number (
nandl)
iValence orbital number (
n,landm)
q
j1andj2pair
p
i1andi2pair
rCPU-rank
Examples:
|
\(D_{\sigma,i_1,i_2}^a\) |
|
|
\(\tilde{n}_\sigma(\mathbf{r})\) |
|
|
\(P_{\sigma \mathbf{k} in}^a\) |
|
|
\(\tilde{\psi}_{\sigma \mathbf{k} n}(\mathbf{r})\) |
|
|
\(\tilde{p}_{\sigma \mathbf{k} i}^a(\mathbf{r}-\mathbf{R}^a)\) |
Domain descriptors¶
GPAW has two different container types for storing one or more functions in a unit cell (wave functions, electron densities, …):
Uniform grids¶
A uniform grid can be created with the UGDesc class:
>>> import numpy as np
>>> from gpaw.core import UGDesc
>>> a = 4.0
>>> n = 20
>>> grid = UGDesc(cell=a * np.eye(3),
... size=(n, n, n))
Given a UGDesc object, one can create
UGArray objects like this
>>> func_R = grid.empty()
>>> func_R.data.shape
(20, 20, 20)
>>> func_R.data[:] = 1.0
>>> grid.zeros((3, 2)).data.shape
(3, 2, 20, 20, 20)
Here are the methods of the UGDesc class:
Size of uniform grid. |
|
Actual size of uniform grid. |
|
Phase factor for block-boundary conditions. |
|
Create new uniform grid description. |
|
Create new UGArray object. |
|
… |
|
Yield views of blocks of data. |
|
Create array of (x, y, z) coordinates. |
|
Create UGAtomCenteredFunctions object. |
|
Create transformer from one grid to another. |
|
Plane wave. |
|
Create UGDesc from grid-spacing. |
|
Create FFTW-plans. |
|
… |
|
Maximum value of ekin so that all 0.5 * G^2 < ekin. |
and the UGArray class:
Create new UniforGridFunctions object of same kind. |
|
Extract x, y values along line. |
|
Return a copy with dtype=complex. |
|
Scatter data from rank-0 to all ranks. |
|
Gather data from all ranks to rank-0. |
|
Do FFT. |
|
Calculate integral over cell of absolute value squared. |
|
Integral of self or self times cc(other). |
|
Convert to UniformGrid with |
|
Multiply by \(exp(ik.r)\). |
|
Interpolate to finer grid. |
|
Restrict to coarser grid. |
|
Add weighted absolute square of data to output array. |
|
Make data symmetric. |
|
Insert random numbers between -0.5 and 0.5 into data. |
|
Calculate moment of data. |
|
Create new scaled UGArray object. |
|
… |
|
… |
Plane waves¶
A set of plane-waves are characterized by a cutoff energy and a uniform grid:
>>> from gpaw.core import PWDesc
>>> pw = PWDesc(ecut=100, cell=grid.cell)
>>> func_G = pw.empty()
>>> func_R.fft(out=func_G)
PWArray(pw=PWDesc(ecut=100 <coefs=1536/1536>, cell=[4.0, 4.0, 4.0], pbc=[True, True, True], comm=0/1, dtype=float64), dims=())
>>> G = pw.reciprocal_vectors()
>>> G.shape
(1536, 3)
>>> G[0]
array([0., 0., 0.])
>>> func_G.data[0]
(1+0j)
>>> func_G.ifft(out=func_R)
UGArray(grid=UGDesc(size=[20, 20, 20], cell=[4.0, 4.0, 4.0], pbc=[True, True, True], comm=0/1, dtype=float64), dims=())
>>> round(func_R.data[0, 0, 0], 15)
1.0
Here are the methods of the PWDesc class:
int([x]) -> integer |
|
Tuple with one element: number of plane waves. |
|
Returns reciprocal lattice vectors, G + k, in xyz coordinates. |
|
Kinetic energy of plane waves. |
|
Create new PlaneWaveExpanions object. |
|
… |
|
Create new plane-wave expansion description. |
|
Return indices into FFT-grid. |
|
Cut out G-vectors with (G+k)^2/2<E_kin. |
|
Paste G-vectors with (G+k)^2/2<E_kin into 3-D FFT grid and |
|
Map from one (distributed) set of plane waves to smaller global set. |
|
Create PlaneWaveAtomCenteredFunctions object. |
and the PWArray class:
Create new PWArray object of same kind. |
|
Create a copy (surprise!). |
|
Sanity check for real-valed PW expansions. |
|
Matrix view of data. |
|
Do inverse FFT(s) to uniform grid(s). |
|
… |
|
Gather coefficients on master. |
|
Gather coefficients from self[r] on rank r. |
|
Scatter data from rank-0 to all ranks. |
|
Scatter all coefficients from rank r to self on other cores. |
|
Integral of self or self time cc(other). |
|
Calculate integral over cell. |
|
Add weighted absolute square of self to output array. |
|
… |
|
Insert random numbers between -0.5 and 0.5 into data. |
|
… |
|
… |
|
… |
Atoms-arrays¶
Block boundary conditions¶
…
Matrix elements¶
>>> psit_nG = pw.zeros(5)
>>> def T(psit_nG):
... """Kinetic energy operator."""
... out = psit_nG.new()
... out.data[:] = psit_nG.desc.ekin_G * psit_nG.data
... return out
>>> H_nn = psit_nG.matrix_elements(psit_nG, function=T)
Same as:
>>> Tpsit_nG = T(psit_nG)
>>> psit_nG.matrix_elements(Tpsit_nG, symmetric=True)
Matrix(float64: 5x5)
but faster.
Atom-centered functions¶
# creates: acf_example.png
import numpy as np
import matplotlib.pyplot as plt
from gpaw.core import UGDesc
alpha = 4.0
rcut = 2.0
l = 0
gauss = (l, rcut, lambda r: (4 * np.pi)**0.5 * np.exp(-alpha * r**2))
grid = UGDesc(cell=[4.0, 2.0, 2.0], size=[40, 20, 20])
pos = [[0.25, 0.5, 0.5], [0.75, 0.5, 0.5]]
acf_aR = grid.atom_centered_functions([[gauss], [gauss]], pos)
coef_as = acf_aR.empty(dims=(2,))
coef_as[0] = [[1], [-1]]
coef_as[1] = [[2], [1]]
print(coef_as.data, coef_as[0])
f_sR = grid.zeros(2)
acf_aR.add_to(f_sR, coef_as)
x = grid.xyz()[:, 10, 10, 0]
y1, y2 = f_sR.data[:, :, 10, 10]
ax = plt.subplot(1, 1, 1)
ax.plot(x, y1, 'o-')
ax.plot(x, y2, 'x-')
# plt.show()
plt.savefig('acf_example.png')
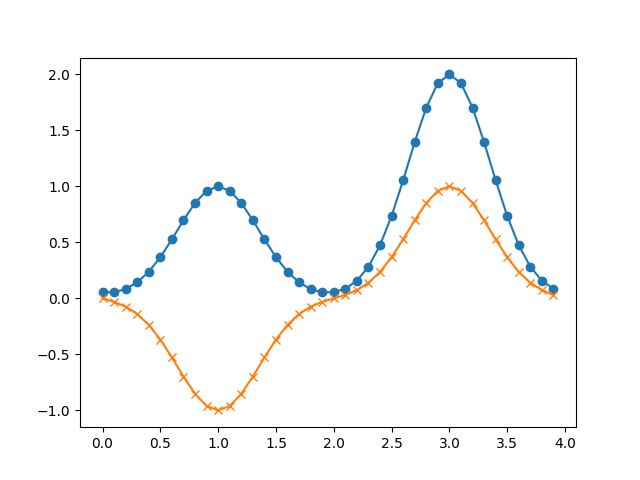
Matrix object¶
Here are the methods of the Matrix class:
Create new matrix of same shape and dtype. |
|
Create a copy. |
|
BLAS matrix-multiplication with other matrix. |
|
Redistribute to other BLACS layout. |
|
Gather the Matrix on the root rank. |
|
Inplace inversion. |
|
In-place inverse of Cholesky decomposition. |
|
Calculate eigenvectors and eigenvalues. |
|
Solve generalized eigenvalue problem. |
|
Inplace complex conjugation. |
|
Add hermitian conjugate to myself. |
|
Fill in upper triangle from lower triangle. |
|
Add list of numbers or single number to diagonal of matrix. |
|
… |
A simple example that we can run with MPI on 4 cores:
from gpaw.core.matrix import Matrix
from gpaw.mpi import world
a = Matrix(5, 5, dist=(world, 2, 2, 2))
a.data[:] = world.rank
print(world.rank, a.data.shape)
Here, we have created a 5x5 Matrix of floats distributed on a 2x2
BLACS grid with a block size of 2 and we then print the shapes of the ndarrays,
which looks like this (in random order):
1 (2, 3)
2 (3, 2)
3 (2, 2)
0 (3, 3)
Let’s create a new matrix b and redistribute
from
a to b:
b = a.new(dist=(None, 1, 1, None))
a.redist(b)
if world.rank == 0:
print(b.array)
This will output:
[[ 0. 0. 2. 2. 0.]
[ 0. 0. 2. 2. 0.]
[ 1. 1. 3. 3. 1.]
[ 1. 1. 3. 3. 1.]
[ 0. 0. 2. 2. 0.]]
Matrix-matrix multiplication works like this:
c = a.multiply(a, opb='T')
API¶
Core¶
- class gpaw.core.UGDesc(*, cell, size, pbc=(True, True, True), zerobc=(False, False, False), kpt=None, comm=MPIComm(size=1, rank=0), decomp=None, dtype=None)[source]¶
Description of 3D uniform grid.
- Parameters:
cell (ArrayLike1D | ArrayLike2D) – Unit cell given as three floats (orthorhombic grid), six floats (three lengths and the angles in degrees) or a 3x3 matrix (units: bohr).
size (ArrayLike1D) – Number of grid points along axes.
pbc – Periodic boundary conditions flag(s).
zerobc – Zero-boundary conditions flag(s). Skip first grid-point (assumed to be zero).
comm (MPIComm) – Communicator for domain-decomposition.
kpt (Vector | None) – K-point for Block-boundary conditions specified in units of the reciprocal cell.
decomp (Sequence[Sequence[int]] | None) – Decomposition of the domain.
dtype – Data-type (float or complex).
- atom_centered_functions(functions, positions, *, qspiral_v=None, atomdist=None, integral=None, cut=False, xp=None)[source]¶
Create UGAtomCenteredFunctions object.
- ekin_max()[source]¶
Maximum value of ekin so that all 0.5 * G^2 < ekin.
In 1D, this will be 0.5*(pi/h)^2 where h is the grid-spacing.
- empty(dims=(), comm=MPIComm(size=1, rank=0), xp=<module 'numpy' from '/scratch/jensj/gpaw-docs/venv/lib/python3.11/site-packages/numpy/__init__.py'>)[source]¶
Create new UGArray object.
- Parameters:
comm (_Communicator) – Distribute dimensions along this communicator.
- fft_plans(flags=0, xp=<module 'numpy' from '/scratch/jensj/gpaw-docs/venv/lib/python3.11/site-packages/numpy/__init__.py'>, dtype=None)[source]¶
Create FFTW-plans.
- classmethod from_cell_and_grid_spacing(cell, grid_spacing, pbc=(True, True, True), kpt=None, comm=MPIComm(size=1, rank=0), dtype=None)[source]¶
Create UGDesc from grid-spacing.
- new(*, kpt=None, dtype=None, comm='inherit', size=None, pbc=None, zerobc=None, decomp=None)[source]¶
Create new uniform grid description.
- property phase_factor_cd¶
Phase factor for block-boundary conditions.
- property size¶
Size of uniform grid.
- class gpaw.core.PWDesc(*, ecut, cell, kpt=None, comm=MPIComm(size=1, rank=0), dtype=None)[source]¶
Description of plane-wave basis.
- Parameters:
ecut (float) – Cutoff energy for kinetic energy of plane waves (units: hartree).
cell (ArrayLike1D | ArrayLike2D) – Unit cell given as three floats (orthorhombic grid), six floats (three lengths and the angles in degrees) or a 3x3 matrix (units: bohr).
comm (MPIComm) – Communicator for distribution of plane-waves.
kpt (Vector | None) – K-point for Block-boundary conditions specified in units of the reciprocal cell.
dtype – Data-type (float or complex).
- atom_centered_functions(functions, positions, *, qspiral_v=None, atomdist=None, integral=None, cut=False, xp=None)[source]¶
Create PlaneWaveAtomCenteredFunctions object.
- empty(dims=(), comm=MPIComm(size=1, rank=0), xp=None)[source]¶
Create new PlaneWaveExpanions object.
- Parameters:
comm (_Communicator) – Distribute dimensions along this communicator.
- map_indices(other)[source]¶
Map from one (distributed) set of plane waves to smaller global set.
Say we have 9 G-vector on two cores:
5 3 4 . 3 4 0 . . 2 0 1 -> rank=0: 2 0 1 rank=1: . . . 8 6 7 . . . 3 1 2
and we want a mapping to these 5 G-vectors:
3 2 0 1 4
On rank=0: the return values are:
[0, 1, 2, 3], [[0, 1, 2, 3], [4]]
and for rank=1:
[1], [[0, 1, 2, 3], [4]]
- new(*, ecut=None, kpt=None, dtype=None, comm='inherit')[source]¶
Create new plane-wave expansion description.
- class gpaw.core.atom_centered_functions.AtomCenteredFunctions(functions, fracpos_ac, atomdist=None, xp=None)[source]¶
- property atomdist¶
- derivative(functions, out=None)[source]¶
Calculate derivatives of integrals with respect to atom positions.
- integrate(functions, out=None)[source]¶
Calculate integrals of atom-centered functions multiplied by functions.
- property layout¶
- class gpaw.core.UGArray(grid, dims=(), comm=MPIComm(size=1, rank=0), data=None, xp=None)[source]¶
Object for storing function(s) on a uniform grid.
- Parameters:
- fft(plan=None, pw=None, out=None)[source]¶
Do FFT.
\[\int d\mathbf{r} e^{i\mathbf{G}\cdot \mathbf{r}} f(\mathbf{r})\]
- fft_restrict(plan1=None, plan2=None, grid=None, out=None)[source]¶
Restrict to coarser grid.
- Parameters:
plan1 (gpaw.fftw.FFTPlans | None) – Plan for FFT.
plan2 (gpaw.fftw.FFTPlans | None) – Plan for inverse FFT.
grid (gpaw.core.uniform_grid.UGDesc | None) – Target grid.
out (gpaw.core.uniform_grid.UGArray | None) – Target UGArray object.
- interpolate(plan1=None, plan2=None, grid=None, out=None)[source]¶
Interpolate to finer grid.
- Parameters:
plan1 (gpaw.fftw.FFTPlans | None) – Plan for FFT (course grid).
plan2 (gpaw.fftw.FFTPlans | None) – Plan for inverse FFT (fine grid).
grid (gpaw.core.uniform_grid.UGDesc | None) – Target grid.
out (gpaw.core.uniform_grid.UGArray | None) – Target UGArray object.
- new(data=None, zeroed=False)[source]¶
Create new UniforGridFunctions object of same kind.
- Parameters:
data – Array to use for storage.
- norm2()[source]¶
Calculate integral over cell of absolute value squared.
\[\int |a(\mathbf{r})|^{2} d\mathbf{r}\]
- class gpaw.core.arrays.DistributedArrays(dims, myshape, comm, domain_comm, data, dv, dtype, xp=None)[source]¶
- abs_square(weights, out)[source]¶
Add weighted absolute square of data to output array.
See also XKCD: 849.
- desc: DomainType¶
- class gpaw.core.atom_arrays.AtomArrays(layout, dims=(), comm=MPIComm(size=1, rank=0), data=None)[source]¶
AtomArrays object.
- Parameters:
layout (AtomArraysLayout) – Layout-description.
comm (MPIComm) – Distribute dimensions along this communicator.
data (np.ndarray | None) – Data array for storage.
- block_diag_multiply(block_diag_matrix_axii, out_ani, index=None)[source]¶
Multiply by block diagonal matrix.
with A, B and C refering to
self,block_diag_matrix_axiiandout_ani:\[\sum^{}_{i} A^{a}_{ni} B^{a}_{ij} \rightarrow C^{a}_{nj}\]If index is not None,
block_diag_matrix_axiimust have an extra dimension: \(B_{ij}^{ax}\) and x=index is used.
- gather(broadcast: Literal[False] = False, copy: bool = False) gpaw.core.atom_arrays.AtomArrays | None[source]¶
- gather(broadcast: Literal[True], copy: bool = False) AtomArrays
Gather all atoms on master.
- new(*, layout=None, data=None, xp=None)[source]¶
Create new AtomArrays object of same kind.
- Parameters:
layout – Layout-description.
data – Array to use for storage.
- to_full()[source]¶
Convert \(N(N+1)/2\) vectors to \(N\times N\) matrices.
>>> a = AtomArraysLayout([6]).empty() >>> a[0][:] = [1, 2, 3, 4, 5, 6] >>> a.to_full()[0] array([[1., 2., 4.], [2., 3., 5.], [4., 5., 6.]])
- class gpaw.core.atom_arrays.AtomArraysLayout(shapes, atomdist=MPIComm(size=1, rank=0), dtype=<class 'float'>, xp=None)[source]¶
Description of layout of atom arrays.
- Parameters:
shapes (Sequence[int | tuple[int, ...]]) – Shapse of arrays - one for each atom.
atomdist (AtomDistribution | MPIComm) – Distribution of atoms.
dtype – Data-type (float or complex).
- empty(dims=(), comm=MPIComm(size=1, rank=0))[source]¶
Create new AtomArrays object.
- Parameters:
comm (_Communicator) – Distribute dimensions along this communicator.
- new(atomdist=None, dtype=None, xp=None)[source]¶
Create new AtomsArrayLayout object with new atomdist.
- class gpaw.core.atom_arrays.AtomDistribution(ranks, comm=MPIComm(size=1, rank=0))[source]¶
Atom-distribution.
- Parameters:
ranks (ArrayLike1D) – List of ranks, one rank per atom.
comm (MPIComm) – MPI-communicator.
- classmethod from_atom_indices(atom_indices, comm=MPIComm(size=1, rank=0), *, natoms=None)[source]¶
Create distribution from atom indices.
>>> AtomDistribution.from_atom_indices([0, 1, 2]).rank_a array([0, 0, 0])
- class gpaw.core.PWArray(pw, dims=(), comm=MPIComm(size=1, rank=0), data=None, xp=None)[source]¶
Object for storing function(s) as a plane-wave expansions.
- Parameters:
- abs_square(weights, out, _slow=False)[source]¶
Add weighted absolute square of self to output array.
With \(a_n(G)\) being self and \(w_n\) the weights:
\[out(\mathbf{r}) \leftarrow out(\mathbf{r}) + \sum^{}_{n} |FFT^{-1} [a_{n} (\mathbf{G})]|^{2} w_{n}\]
- gather_all(out)[source]¶
Gather coefficients from self[r] on rank r.
On rank r, an array of all G-vector coefficients will be returned. These will be gathered from self[r] on all the cores.
- ifft(*, plan=None, grid=None, out=None, periodic=False)[source]¶
Do inverse FFT(s) to uniform grid(s).
- Parameters:
plan – Plan for inverse FFT.
grid – Target grid.
out – Target UGArray object.
- new(data=None)[source]¶
Create new PWArray object of same kind.
- Parameters:
data – Array to use for storage.
- norm2(kind='normal')[source]¶
Calculate integral over cell.
For kind=’normal’ we calculate:
\[\int |a(\mathbf{r})|^{2} d\mathbf{r} = \sum^{}_{G} |c_{G} |^{2} V,\]where V is the volume of the unit cell.
And for kind=’kinetic’:
\[\frac{1}{2} \sum^{}_{G} |c_{G} |^{2} G^{2} V,\]
- class gpaw.core.matrix.Matrix(M, N, dtype=None, data=None, dist=None, xp=None)[source]¶
Matrix object.
- Parameters:
M (int) – Rows.
N (int) – Columns.
dtype – Data type (float or complex).
dist (MatrixDistribution | tuple | None) – BLACS distribution given as (communicator, rows, columns, blocksize) tuple. Default is None meaning no distribution.
data (ArrayLike2D | None) – Numpy ndarray to use for storage. By default, a new ndarray will be allocated.
- eigh(S=None, *, cc=False, scalapack=(None, 1, 1, None), limit=None)[source]¶
Calculate eigenvectors and eigenvalues.
Matrix must be symmetric/hermitian and stored in lower half. If
Sis given, solve a generalized eigenvalue problem.
- eighg(L, comm2=MPIComm(size=1, rank=0))[source]¶
Solve generalized eigenvalue problem.
With \(H\) being self, we solve for the eigenvectors \(C\) and the eigenvalues \(Λ\) (a diagonal matrix):
\[HC = SCΛ,\]where \(L\) is a lower triangle matrix such that:
\[LSL^{†} = 1\cdot\]The solution has these three steps:
\[\tilde{H} = LHL^{†} , \tilde{H}\tilde{C} = \tilde{C}Λ, C = L^{†} \tilde{C}\cdot\]Note that \(H\) must be the full matrix not just half of it!
- gather(root=0, broadcast=False)[source]¶
Gather the Matrix on the root rank.
Returns a new Matrix distributed so that all data is on the root rank
- invcholesky()[source]¶
In-place inverse of Cholesky decomposition.
Calculate a lower triangle matrix \(L\) where:
\[LSL^{†} = 1,\]and \(S\) is self. Only the lower part of \(S\) is used.
>>> S = Matrix(2, 2, data=[[1.0, np.nan], ... [0.1, 1.0]]) >>> S.invcholesky() >>> S.data array([[ 1. , -0. ], [-0.10050378, 1.00503782]])
- multiply(other, alpha=1.0, opa='N', opb='N', out=None, beta=0.0, symmetric=False)[source]¶
BLAS matrix-multiplication with other matrix.
DFT¶
- class gpaw.new.calculation.DFTCalculation(state, setups, scf_loop, pot_calc, log)[source]¶
- converge(convergence=None, maxiter=None, steps=99999999999999999, calculate_forces=None)[source]¶
Converge to self-consistent solution of Kohn-Sham equation.
- class gpaw.new.calculation.DFTState(ibzwfs, density, potential)[source]¶
State of a Kohn-Sham calculation.
- class gpaw.new.density.Density(nt_sR, taut_sR, D_asii, charge, delta_aiiL, delta0_a, N0_aii, n_aj, l_aj, nct_aX, tauct_aX)[source]¶
- classmethod from_superposition(*, grid, nct_aX, tauct_aX, atomdist, setups, basis_set, magmom_av, ncomponents, charge=0.0, hund=False, mgga=False)[source]¶
- property nct_R¶
- property tauct_R¶
- gpaw.new.builder.builder(atoms, params, comm=None)[source]¶
Create DFT-components builder.
pw
lcao
fd
tb
atom
- class gpaw.new.ibzwfs.IBZWaveFunctions(ibz, *, nelectrons, ncomponents, wfs_qs, kpt_comm=MPIComm(size=1, rank=0), kpt_band_comm=MPIComm(size=1, rank=0), comm=MPIComm(size=1, rank=0))[source]¶
Collection of wave function objects for k-points in the IBZ.
- classmethod create(*, ibz, nelectrons, ncomponents, create_wfs_func, kpt_comm=MPIComm(size=1, rank=0), kpt_band_comm=MPIComm(size=1, rank=0), comm=MPIComm(size=1, rank=0))[source]¶
Collection of wave function objects for k-points in the IBZ.
- get_all_electron_wave_function(band, kpt=0, spin=0, grid_spacing=0.05, skip_paw_correction=False)[source]¶
- get_max_shape(global_shape=False)[source]¶
Find the largest wave function array shape.
For a PW-calculation, this shape could depend on k-point.
- property mode¶
- class gpaw.new.pot_calc.PotentialCalculator(xc, poisson_solver, setups, *, fracpos_ac, external_potential=None, soc=False)[source]¶
- class gpaw.new.scf.SCFLoop(hamiltonian, occ_calc, eigensolver, mixer, comm, convergence, maxiter)[source]¶
- class gpaw.new.pwfd.wave_functions.PWFDWaveFunctions(psit_nX, *, spin, q, k, setups, fracpos_ac, atomdist, weight=1.0, ncomponents=1, qspiral_v=None)[source]¶
- property P_ani: AtomArrays¶
PAW projections.
\[\langle \tilde{p}^{a}_{i}|\tilde{ψ}_{n} \rangle\]
- dipole_matrix_elements()[source]¶
Calculate dipole matrix-elements.
\[\mathbf{μ}_{mn} = \int d\mathbf{r} \tilde{𝜓}_{m} \tilde{𝜓}_{n} \mathbf{r} + \sum^{}_{aij} P^{a}_{im} P^{a}_{jn} Δ\mathbf{μ}^{a}_{ij}\]- Returns:
matrix elements in atomic units.
- Return type:
Array3D
- orthonormalize(work_array_nX=None)[source]¶
Orthonormalize wave functions.
Computes the overlap matrix:
\[S_{mn} = \int \tilde{ψ}_{m}(\mathbf{r})^{*} \tilde{ψ}_{n}(\mathbf{r}) d\mathbf{r} + \sum^{}_{aij} (P^{a}_{im} )^{*} P^{a}_{jn} ΔS^{a}_{ij}\]With \(LSL^\dagger=1\), we update the wave functions and projections inplace like this:
\[Ψ_{m} \leftarrow \sum^{}_{n} L^{*}_{mn} Ψ_{n} ,\]and:
\[P^{a}_{mi} \leftarrow \sum^{}_{n} L^{*}_{mn} P^{a}_{ni} \cdot\]
- property pt_aiX: AtomCenteredFunctions¶
PAW projector functions.
\[\tilde{p}^{a}_{i} (\mathbf{r})\]
- class gpaw.new.ase_interface.ASECalculator(params, *, log, dft=None, atoms=None)[source]¶
This is the ASE-calculator frontend for doing a GPAW calculation.
- calculate_property(atoms, prop)[source]¶
Calculate (if not already calculated) a property.
The
propstring must be one ofenergy
forces
stress
magmom
magmoms
dipole
- property density¶
- property dft: DFTCalculation¶
- dos(soc=False, theta=0.0, phi=0.0, shift_fermi_level=True)[source]¶
Create DOS-calculator.
Default is to
shift_fermi_levelto 0.0 eV. Forsoc=True, angles can be given in degrees.
- property hamiltonian¶
- iconverge(atoms)[source]¶
Iterate to self-consistent solution.
Will also calculate “cheap” properties: energy, magnetic moments and dipole moment.
- implemented_properties = ['energy', 'free_energy', 'forces', 'stress', 'dipole', 'magmom', 'magmoms']¶
- property initialized¶
- name = 'gpaw'¶
- property parameters¶
- property results¶
- property setups¶
- property spos_ac¶
- property symmetry¶
- property wfs¶
- property world¶
FFTW¶
Python wrapper for FFTW3 library¶
- class gpaw.fftw.FFTWPlans(size_c, dtype, flags=0)[source]¶
FFTW3 3d transforms.
- class gpaw.fftw.NumpyFFTPlans(size_c, dtype, empty=<function empty>)[source]¶
Numpy fallback.
- gpaw.fftw.check_fft_size(n, factors=[2, 3, 5, 7])[source]¶
Check if n is an efficient fft size.
Efficient means that n can be factored into small primes (2, 3, 5, 7).
>>> check_fft_size(17) False >>> check_fft_size(18) True
- gpaw.fftw.create_plans(size_c, dtype, flags=0, xp=<module 'numpy' from '/scratch/jensj/gpaw-docs/venv/lib/python3.11/site-packages/numpy/__init__.py'>)[source]¶
Create plan-objects for FFT and inverse FFT.
- gpaw.fftw.empty(shape, dtype=<class 'float'>)[source]¶
numpy.empty() equivalent with 16 byte alignment.
BLAS¶
- gpaw.utilities.blas.mmm(alpha, a, opa, b, opb, beta, c)[source]¶
Matrix-matrix multiplication using dgemm or zgemm.
For opa=’N’ and opb=’N’, we have:
\[c \leftarrow αab + βc\cdot\]Use ‘T’ to transpose matrices and ‘C’ to transpose and complex conjugate matrices.
- gpaw.utilities.blas.rk(alpha, a, beta, c, trans='c')[source]¶
Rank-k update of a matrix.
For
trans='c'the following operation is performed:\[c \leftarrow αaa^{†} + βc,\]and for
trans='t'we get:\[c \leftarrow αa^{†} a + βc\]If the
aarray has more than 2 dimensions then the 2., 3., … axes are combined.Only the lower triangle of
cwill contain sensible numbers.